Posted by star
on 2019-10-20 18:33:21
Hits:769
Researchers at the Case Western Reserve University School of Medicine and the Cleveland University Hospital Medical Center have discovered a way to enhance kidney self-repair. This is a way to save the damaged kidney by reprogramming the self metabolism.
The glycolysis process converts glucose into energy, and new research suggests that when tissue is damaged, the body switches this pathway to repair damaged cells. When cells break down fat, sugar, and protein into glucose, all three substances are first converted to intermediates and then into the mitochondria. Researchers report that when a tissue is injured, such as a kidney tissue, the body converts glucose into another new molecule and then takes the cell toward repair.
The key substance controlling the path of glucose is called PKM2 protein. Inactivation of PKM2 resulted in a significant increase in cell repair and a reduction in energy production. After the disease or injury, the animal attempts to disable the PKM2 protein in order to transfer the glucose to the repair pathway. In the experiment, after the inhibition of amplification, the kidney damage of mice has a more obvious repair effect.
One of the key molecules in tissue repair, nitric oxide (NO), is a well-known umbrella for kidneys and other tissues. It is also an active component of the heart drug nitroglycerin, which has been thought to play a role in dilating blood vessels. However, in the new study, the team found that NO adheres to coenzyme A, a key molecule involved in glycolysis and energy production, and binds and transports NO to many different proteins, including PKM2 to shut them down. This is the idea that kidney cells decide to “repair” or “capacity”.
In addition to discovering that the addition of NO to PKM2 can activate repair, the team found that another protein called AKR1A1 is responsible for removing NO from PKM2 and re-activating a powerful energy production process. After the healing......
Posted by star
on 2019-10-17 18:53:45
Hits:646
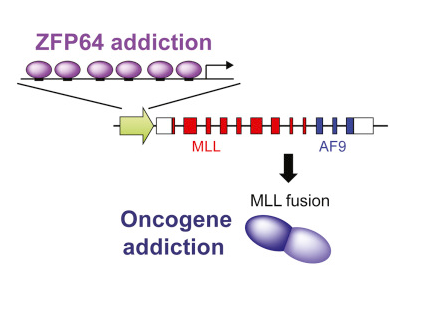
About 5% of cases of acute leukemia are diagnosed as mixed leukemia (MLL), which is common in infants. The cause is a gene called MLL, which produces a protein of the same name to promote new cell growth. In most cases, MLL is properly regulated by the body and does not cause disease. Unfortunately, in some cases, MLL incorrectly produces MLL fusion proteins.
Researchers from CSHL found that the MLL gene encoding the MLL fusion protein that causes a series of troubles is easily activated by ZFP64, a special protein.
ZFP64 is very common in healthy people, but no one has considered it to be a causative factor in any type of leukemia. Further testing showed that the production of the MLL fusion protein is completely dependent on this unique, unobtrusive protein.
The critical function of ZFP64 in leukemia is to maintain MLL expression via binding to the MLL promoter, which is the most enriched location of ZFP64 occupancy in the human genome. The specificity of ZFP64 for MLL is accounted for by an exceptional density of ZFP64 motifs embedded within the MLL promoter. These findings demonstrate how a sequence anomaly of an oncogene promoter can impose a transcriptional addiction in cancer.
"The ZFP64 deletion is a highly desirable drug target because it completely and specifically inhibits MLL fusion protein production and leukemia cascade growth," the investigators said.
EIAAB SCIENCE INC, WUHAN has developed ZFP64 protein, antibody and ELISA kit.
Posted by star
on 2019-10-16 19:19:55
Hits:490
Branched or lobed DNAs with single-stranded ends are formed during DNA replication and DNA repair in healthy individuals. These single-stranded flap DNAs can be excised by an enzyme such as FEN1 to repair the integrity of the double-stranded state of the DNA.
Higher levels of FEN1 have been observed in cancer cells with worse clinical outcomes, and these cells are more dependent on this repair enzyme in the absence of other supporting DNA damage response proteins. These cells are sensitive and will die when FEN1 is blocked - synthetically lethal. Preventing the action of FEN1 such enzymes may be a way to selectively kill these cancer cells.
An interdisciplinary team of scientists from the University of Sheffield and AstraZeneca Pharmaceuticals confirmed in detail the combination of inhibitors that block and block FEN1 to provide clues as to how "the function of the protein is blocked."
They present the first crystal structure of inhibitor-bound hFEN1 and show a cyclic N-hydroxyurea bound in the active site coordinated to two magnesium ions. Three such compounds had similar IC50 values but differed subtly in mode of action. One had comparable affinity for protein and protein–substrate complex and prevented reaction by binding to active site catalytic metal ions, blocking the unpairing of substrate DNA necessary for reaction. Other compounds were more competitive with substrate. Cellular thermal shift data showed engagement of both inhibitor types with hFEN1 in cells with activation of the DNA damage response evident upon treatment. However, cellular EC50s were significantly higher than in vitro inhibition constants.
The researchers said: "The results of this study have indeed deepened our understanding of how 'a group of specific compounds previously identified as FEN1 inhibitors work.' This insight can help us develop methods to block nucleases, nucleases are A family of important enzymes in cellular DNA damage response, AstraZeneca's st......
Posted by star
on 2019-10-15 19:07:34
Hits:578
The consciousness of the brain is the basis of all human activities, if the brain activity disorder or stop, human life will be seriously threatened. Alzheimer's disease (AD) is a relatively common brain disease characterized by progressive memory loss and cognitive impairment. It is not yet clear what brain activity causes Alzheimer's, but the production and accumulation of beta-amyloid has long been thought to be an important factor.
Aβis encoded by the APP produced through A series of hydrolase enzyme, the APP is Aβprecursor protein, full name is the amyloid precursor protein, APP across the membrane in the process of production there are 2 hydrolysis ways, which does not produce AD the starch source way and generate Aβsource of starch way, in the first article in hydrolysis pathways, APP proteolytic and secretion after α-secretes enzyme action, produce soluble α- APPs and released into the extracellular, leave the APP alpha CTF at the same time, this process destroys A complete structure of beta, no neurotoxicity, this process is known as the amyloid production means, under normal circumstances, This metabolic pathway is dominant. Aβis produced by the starch source pathway. APP, under the action ofβ-secretase, secretes soluble beta-apps and c-terminal fragments (β-CTF) from the n-terminal of Aβregion, and then continues to cleave by γ-secretase, producing a series of Aβand AICD of different lengths. In this process, if the balance between the production and clearance of Aβis broken, abnormal deposition of Aβ in the cerebral cortex will be caused, which will lead to synaptic damage and degeneration of neurons.
So what exactly is the relationship between Aβand AD? The pathogenesis of AD is very complex. At present, there is no clear explanation for the pathogenesis of AD, and many theories are in the hypothesis stage. The dominant hypothesis is the Aβ hypothesis. This hypoth......
Posted by star
on 2019-10-14 18:31:09
Hits:560
The human digestive tract follows a pattern of digesting food and nutrients while awake and replenishing senescent cells while asleep. But shift work and jet lag can disrupt the body clock, which has been linked to increased risks of intestinal infections, obesity, inflammatory bowel disease and colorectal cancer.
Now, researchers at the University of California School of medicine have discovered an immune cell that helps keep the gut healthy. These cells, called type 3 innate lymphoid cells (ILC3), are responsible for keeping the gut functioning normally and healthily. The researchers found that so-called clock genes were highly active in these cells, which produced immune molecules closely related to clock gene activity. When researchers knocked out a key clock gene in mice, the mice were unable to produce a subset of ILC3 cells and had difficulty controlling bacterial infections in their guts.
The findings could help explain why disruptions in circadian rhythms often lead to gastrointestinal problems. In addition, they point out, the clock's genes can affect immune cells and help counteract the negative effects of sleep irregularities associated with bowel disease.
"High strength work, insomnia, chronic sleep deprivation on the destruction of the circadian rhythm is more and more obvious, and have harmful effects on human health, but we still don't know why interrupted sleep can cause these problems," the researchers Bruce r. Stevens said, "now we have found that circadian rhythms directly affects the function of immune cells in the gut, which could help explain some health problems, such as inflammatory bowel disease."
ILC3 cells maintain the balance in the gut. They also produce immune molecules that help the gut immune system avoid overreacting to harmless microbes and food particles, while maintaining its ability to fight disease-causing microbes.
The researchers studied ILC3 cells taken from the intestinal tract of mice every six ......
