Posted by star
on 2019-09-10 19:10:59
Hits:523
A new study from the University of Texas shows that older adults who have never participated in a sustained exercise program have the same ability to build muscle mass as athletes of the same age who train over time.
Studies show that even people who don't exercise at all can benefit from resistance training such as weight training.
Researchers at the University of Texas School of sports and exercise science compared the muscle-building abilities of two groups of older men. The first group were athletes who were lifelong exercisers; the second group was healthy individuals of a similar age who had never had exercise training.
Each participant will receive an isotopic tracer and then participate in an exercise program, including weight training on an exercise machine. The researchers examined muscle biopsies taken 48 hours before and after exercise for signs of how the muscles responded to exercise. Isotope tracers show how proteins develop in muscles.
The researchers had expected athletes to have more muscular capacity than those who did not train. In fact, the results showed that both groups had the same muscular capacity for exercise.
"Our research clearly shows that if you are not a regular exerciser, you can benefit from exercise whenever you start," said Dr. W. Larry Kenney of the University of Texas. Clearly, a long-term commitment to physical fitness and exercise is the best way to achieve overall health, or to start exercising later, which can also help stave off weakness and muscle weakness in older people.

Posted by star
on 2019-09-09 19:13:41
Hits:559
The researchers observed protein aggregates in the nerve tissue of Parkinson's patients, composed of the protein alpha-synuclein monomers that assemble into so-called amyloid fibrils. Similar deposits have also been found in other neurodegenerative diseases such as Alzheimer's. So researchers think these fibrils are responsible for the disease, and to test that possibility, researchers are looking for ways to stop fibrils from forming.
Professor William h.Crocker describes a class of engineered binding proteins called beta-wrapins that prevent alpha-synaptic nucleonuclear proteins from clumping together. "We studied the function of beta-wrapins and how they disrupt the process of alpha-synaptic nuclein aggregation," says William.

First, the researchers found that beta-wrapins prevented the new alpha-synuclein monomer from lengthening amyloid fibrils. Because β-wrapins capture monomers and form chemical complexes with them. In addition, the researchers studied the role of β-wrapins not only in test tubes, but also in cell culture and animal models. The researchers treated diseased flies with beta-encapsulating proteins and showed significantly improved motor skills in climbing tests.
Posted by star
on 2019-09-08 23:17:34
Hits:519
As you may know, in the human lung lined with hair-like protrusions, we are going to these bumps are called cilia movement, the cilia is composed of some tiny tubes, usually appear on the surface of certain cells or tissues, for example, in our there is movement in the nose and respiratory cilia, these cilia movement will move from side to side, remove microorganisms, mucus, and death cells in the respiratory system, etc. However, there may be a more important subclass of motile cilia, called ciliary. ciliary are found in almost all cells in the body, but they are not always present.
The functional structure of these cilia is very important. Interfering with their formation or function can lead to cilia diseases, which often cripple and endanger the life of patients and affect the functions of multiple organs in the body. Why does the failure of such a small organelle cause a variety of disease symptoms? The researchers found that cilia are packed with proteins that detect signals from other cells and their surroundings, which are then sent to the nucleus to activate the body's reactions, which are crucial for regulating the body's signaling pathways. And these signaling pathways are also associated with cancer, so it is particularly important to study the relationship between cilia and cancer.
The researchers found that in many cancers, there was a significant lack of ciliary structure in cancer cells compared to surrounding healthy cells. This may be because the absence of cilia is a response to cancer, interfering with the regulation of normal cells.
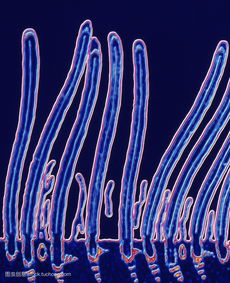
Melanoma is a malignant skin cancer, and some melanoma cells will express higher levels of EZH2 protein, which will inhibit the expression of cilia gene so that melanoma has less cilia structure, and the absence of cilia can activat......
Posted by star
on 2019-09-05 18:53:14
Hits:559
Spitzoid melanoma is a specific morphologic variant of melanoma that most commonly affects children and adolescents, and ranges on the spectrum of malignancy from low grade to overtly malignant. These tumors are generally driven by fusions of ALK, RET, NTRK1/3, MET, ROS1 and BRAF1,2. However, in approximately 50% of cases no genetic driver has been established.
A team led by St. Jude Children's Research Hospital has demonstrated the potential to use clinical genome sequencing to treat Spitzoid-type melanoma.
The study involved an 11-year-old boy with locally recurrent spitzoid melanoma who had previously tried traditional treatments but had poor results. The researchers collected clinical samples, performed whole-genome, exome, and RNA sequencing, and found MAP3K8 gene fusion, which encodes a serine-threonine kinase that activates MEK3.
The researchers then used RNA sequencing to evaluate 51 samples from another 49 patients with spitzoid-like melanoma. They found a fusion associated with MAP3K8 in almost one-third of cases. They pointed out that each fusion involves the substitution or truncation of self-inhibiting exons in MAP3K8 .
"The disruption of MAP3K8 leads to deletion or replacement of exons, which is the most common change in 49 cases," the authors said. "Rearrangement has been found in atypical Spitz tumors and spitzoid melanoma cases, as with other mutations or fusions, these rearrangements may also occur in benign nevi."
Based on these findings, the authors note that "MAP3K8 rearrangement is the most common genetic event in spitzoid-type melanoma and is also present in adult melanoma and may be suitable for the treatment of MEK inhibitors."
EIAAB SCIENCE INC, WUHAN has developed MAP3K8 protein, antibody and ELISA kit.
Welcome scientific research workers to choose and purchase.

Posted by star
on 2019-09-05 00:54:06
Hits:509
Parkinson's disease (PD) is a common degenerative disease of the nervous system, and its clinical symptoms are mainly characterized by rigidity and tremor. Currently, approximately 6 million people worldwide suffer from Parkinson's disease.
The researchers from Oxford used high-resolution, single-cell transcriptomic analyses of iPSC-derived dopamine neurons carrying the GBA-N370S PD risk variant, they identified a progressive axis of gene expression variation leading to endoplasmic reticulum stress. Pseudotime analysis of genes differentially expressed (DE) along this axis identified the transcriptional repressor histone deacetylase 4 (HDAC4) as an upstream regulator of disease progression. HDAC4 was mislocalized to the nucleus in PD iPSC-derived dopamine neurons and repressed genes early in the disease axis, leading to late deficits in protein homeostasis. Treatment of iPSC-derived dopamine neurons with HDAC4-modulating compounds upregulated genes early in the DE axis and corrected PD-related cellular phenotypes.Our study demonstrates how single-cell transcriptomics can exploit cellular heterogeneity to reveal disease mechanisms and identify therapeutic targets.
They also added that many of the differentially expressed genes were down-regulated by histone deacetylase 4 (HDAC4), suggesting that targeting it may affect disease processes. They found that HDAC4 is incorrectly localized on the nucleus of patient iPSC-differentiated dopamine neurons, inhibiting gene expression at an early stage, leading to defects in late protein balance.
The researchers then used four compounds that target HDAC4 to treat dopamine neurons in patients. They reported that these compounds reversed the decline in expression of early disease-associated genes, and that further processing later limited the expression of endoplasmic reticulum stress genes.
"We found that the disease process is a dynamic event and determined that HDAC4 is a key regulator of early molecular ......
