Posted by star
on 2019-11-13 23:24:07
Hits:2632
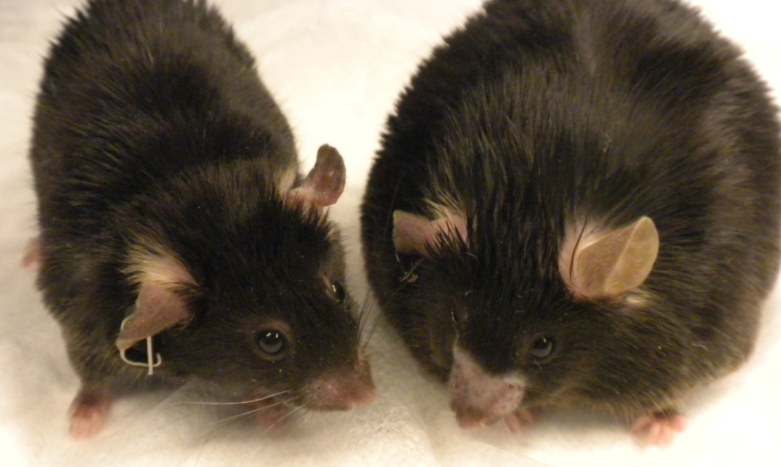
Obesity and obesity-related diseases, such as type 2 diabetes and liver disease, constitute a major burden on global health. Researchers from the Cincinnati Children's Hospital Medical Center reported that blocking RNA silencing in the liver of mice allowed animals to Stay away from obesity and diabetes.
The team thoroughly screened and analyzed the activity of liver genes and their molecular targets (such as key proteins such as AMPK) and found that genetically deleted Argonaute 2 (Ago2) in the liver of mice, which affects energy metabolism by controlling RNA silencing. . When it silences RNA in the liver, metabolism and the ability of the liver to treat a high-fat diet are reduced.
Deletion of Ago2 in the liver is not toxic to animals, but it does stabilize energy metabolism. "Although this is still a basic study, we believe that this may be important for obesity-related chronic metabolic disorders such as diabetes and fatty liver," the researchers said. “We can continue to explore new ways to change the energy balance and regulation of obesity.”
"The role of Ago2 is the "joining point" between liver protein translation, energy production depletion, and AMPK activity. Functional disruption of these events is a common feature of obesity and its related diseases." The researchers said
The scientists pointed out that this is an early study, they need to further verify in the laboratory model, and finally develop a practical drug to inhibit Ago2 in the clinical environment.
EIAAB SCIENCE INC, WUHAN has developed Ago2 protein, antibody and ELISA kit.
Posted by star
on 2019-11-10 19:37:47
Hits:876
A study led by Georgia State University showed beta-hydroxybutyrate produced during fasting or calorie restriction. It has an anti-aging effect on the vascular system, which can reduce and alleviate blood vessel-related diseases such as cardiovascular disease.
A small molecule of β-hydroxybutyrate is a kind of ketone body (or a water-soluble molecule containing a ketone group) which is produced from fatty acids in the liver under the following condtions: low food intake and carbohydrates, starvation and long-term intense exercise. β-hydroxybutyric acid can delay vascular aging by endothelial cells arranged on the inner surface of blood vessels and lymphatic vessels. It can block a type of aging called senescence or cellular aging, where cells have function but exit the cell cycle.
The aging cells can no longer divide. The researchers found that beta-hydroxybutyrate promotes cell division and prevents these cells from aging. Because this molecule is produced during calorie restriction or fasting, when people overeating or becoming obese, the molecule may be inhibited, which will accelerate aging.
In addition, the researchers found that when β-hydroxybutyric acid binds to specific RNA-binding proteins, it increases the activity of the transcription factor Oct4 (Octamer-binding transcriptional factor) in mouse vascular smooth muscle and endothelial cells. OCT4 increases a key factor against DNA damage-induced aging and keeps blood vessels younger.
This stem cell factor (OCT4) may be a drug or pharmacological target for slowing or preventing aging. If the vascular system becomes younger, it is less likely to suffer from cardiovascular disease, Alzheimer's and cancer, as all of these diseases are associated with aging.
In the future, researchers hope to target aging cells in order to eliminate them and restore the vascular system to prevent cardiovascular disease.
EIAAB SCIENCE INC, WUHAN has developed
Posted by star
on 2019-10-31 18:44:42
Hits:856
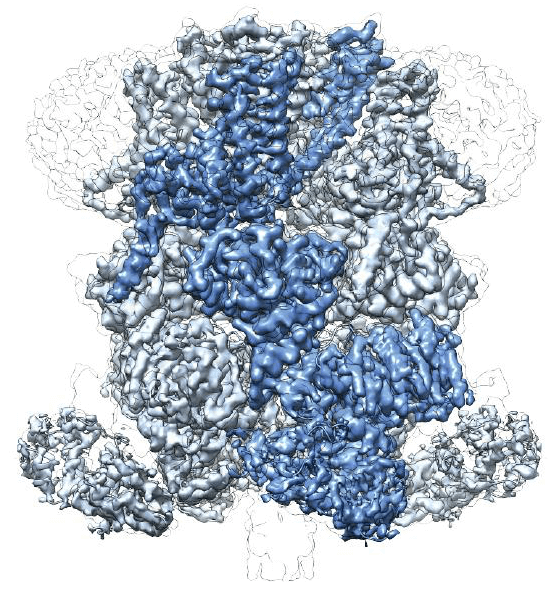
Scientists at the Van Andel Institute (VARI) have for the first time revealed the atomic structure of the protein TRPM2, which regulates core body temperature, mediates immune responses, and apoptosis, programmed cell death.
TRPM2 is part of the TRP superfamily, which mediates sensory stimuli such as pain, stress, vision, temperature, and taste. They are widely known as ion channel proteins, nested within the cell membrane, and act as chemistry into and out of cells. The gatekeeper of the signal. The eight proteins that make up the TRPM superfamily are part of this broader group.
TRPM2 protein may be a promising drug target for the treatment of oxidative stress-related neurological diseases (Alzheimer's and bipolar disorder, etc.) caused by chemical imbalances.
The atomic image of TRPM2 depicts a bell-like structure with a transmembrane domain on the shoulder of the clock and an extended NUDT9-H domain at the bell. In addition, the study also identified a new drug-binding site for ADPR, a signaling molecule involved in oxidative stress and metabolism, a finding that reversed the previously identified view that TRPM2 binds to ADPR in the NUDT9-H domain. The exact drug targets provide valuable details for designing treatments for body temperature-related diseases and preventing neuronal death during neurodegenerative diseases.
"Although we know that TRPM2 is a component of many important life processes and potential drug targets, we are not sure what it looks like or how it works," the researchers said. “Today's discovery has changed all of this and helped to understand this extremely important molecule more fully.”
EIAAB SCIENCE INC, WUHAN has developed TRPM2 protein, antibody and ELISA kit.
Welcome scientific research workers to choose and purchase.
Posted by star
on 2019-10-29 18:25:21
Hits:1051
Researchers at the Salk Institute in the United States have determined how certain cells in the eye handle ambient light and reset our biological clock. Late at night, when these cells are exposed to artificial light, our biological clocks can become confusing, leading to a range of health problems.
The innermost layer of our retina contains a group of light-sensitive cells. When these cells are exposed to light, a protein called melanopins is continuously regenerated, sending levels of ambient light directly to the brain to regulate consciousness, sleep and alertness. It plays a key role in synchronizing our biological clocks, and a 10-minute exposure to intense light suppresses melatonin, which regulates sleep.
In this study, researchers used molecular tools to initiate the production of melanopsin in mouse retinal cells. They found that some cells maintained their ability to respond lightly when exposed to repeated long-pulsed light, while others became insensitive.
The traditional wisdom is that the arrestin should stop the photoreaction of the cells within a few seconds of the light being on. However, the researchers were surprised to find that the inhibitory protein is actually required for melanin to respond to long-term light exposure.
In mice lacking inhibitory proteins (β-arrestin 1 and β-arrestin 2), retinal cells producing melanops were unable to maintain their sensitivity to light under prolonged exposure. This result just indicates that the inhibitory protein contributes to the regeneration of melanopsin in retinal cells.
"Our results indicate that the two inhibitory proteins achieve the regeneration of melanopsin in a unique way. One inhibitory protein plays a traditional role in stopping the photoreactive response, while the other inhibits the retinal sensitization cofactor of melanopsin. When these two steps alternate rapidly, the cells seem to be constantly responding to light," the researchers said.
To be......
Posted by star
on 2019-10-22 19:16:22
Hits:1313
Researchers at the University of Texas Southwestern Medical Center have discovered how DNA can trigger immune and autoimmune responses from within cells by discovering the DNA sensing enzyme cGAS.
The NLRP3 protein is the basis for inflammation of a group of autoinflammatory diseases called cryopyrin protein-associated periodic syndrome (CAPS), including familial cold urticaria (FCAS), gout and an Alzheimer's disease and disease-related brain cell inflammation. In order to cope with excessive harmful substances, including toxins and cholesterol crystals, the inflammatory body initiates the path of inflammatory cell death. Inflammatory bodies also increase the body's immune system substances, such as interleukins, which contribute to the body's immune response.
"A long-standing problem in this area is how NLRP3 is activated by many different drugs that do not appear to have any chemical or structural similarities," the researchers said.
They discovered a previously unknown structural change in NLRP3 in the cell through imaging and genetic methods. They found that different stimuli caused an organelle called the reverse Golgi network to split into giant vesicles or fluid vesicles. These vesicles contain a special lipid component (PI4P) that binds to specific regions of NLRP3, and their binding triggers a series of events that lead to inflammatory body activation.
"The uniqueness of the NLRP3 inflammatory corpuscle is that it can be stimulated by a large number of stimuli," the researchers said. "This study found that the NLRP3 inflammatory body does not directly recognize harmful substances, but rather detects the structural changes caused by a series of different compounds that cause cell damage. In fact, the activation of NLRP3 is reminiscent of the plant's 'guard model', a model that counters threats by detecting host targets that have been altered, so-called pathogen induction."
"The NLRP3 indirectly perceives various pathogens and dang......
